The mean donor-acceptor distances in protein secondary structure elements are close to 3.0 Å, as are those between bases in Watson-Crick pairing (Jeffrey[1], pp. 191, 200). Jeffrey[1] (page 12) categorizes hbonds with donor-acceptor distances of 2.2-2.5 Å as "strong, mostly covalent", 2.5-3.2 Å as "moderate, mostly electrostatic", and 3.2-4.0 Å as "weak, electrostatic". Energies are given as 40-14, 15-4, and <4 kcal/mol respectively. Most hbonds in proteins are in the moderate category. Strong hbonds require moieties or conditions that are rare within proteins. The hydrogen atoms in moderate hbonds often do not lie on the straight line connecting the donor to acceptor, so donor-acceptor distance slightly underestimates the length of the hbond (Jeffrey[1], p. 14). http://proteopedia.org/wiki/index.php/Hydrogen_bonds All kinds of biological behavior are taken place in aqueous state, thus the role of water in these processes can not be excluded. This page will be mainly talking about how the features of water especially hydrogen bond network influence the behavior of large biomolecules. First thing I can think of is to study the water network, I believe all kinds of functions must be performed through the inside factor which is biomolecules with the help of outside factor which is the solvent and other complementary molecules, and they need to work together to realize these functions. the main relationship between biomolecules and waters are hydrogen bonds. When we talk about hydrogen bond, we don't think of it as exactly non-covalent bond, they have some part of genes from covalent bond, look at the picture below(1): 
If you look the A, the hydrogen bonds connected these two molecules are slightly bright, or it will be totally dark like C. Yes, the hydrogen bond are transient, it cannot sustain long time like regular covalent bond, but average effect of this intermittent hydrogen bond will be more like a weak covalent bond.
There are two definitions of hbond in simulation:(i) energetic definition, which consider two molecules to be bonded if their oxygen-oxygen separation is less than 3.5A and their interaction energy is less than a threshold energy 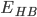 .(ii)geometric definition,which use the same distance criterion but no energetic condition, instead requiring that the O-H
.(ii)geometric definition,which use the same distance criterion but no energetic condition, instead requiring that the O-H O angle between two molecules much be less than a threshold angle
O angle between two molecules much be less than a threshold angle 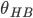 .(2)
.(2)
First let's look at the lifetime of hbond. The relaxation time for water dipole reorientation at room temperature is about 10ps, the average hydrogen bond lifetime will roughly agree with (or smaller than)this value because reorientation is impossible without breaking at least one hbond. The picture below is calculation of average hbond lifetime (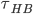 ) from reference 2.
) from reference 2.
(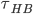 is the mean of the distribution of hbond lifetime P(t), which measures the probability that an initially bonded pair remains bonded at all times up to time t, and breaks at t. P(t) is obtained from simulations by building a histogram of hbond lifetimes for each configuration,
is the mean of the distribution of hbond lifetime P(t), which measures the probability that an initially bonded pair remains bonded at all times up to time t, and breaks at t. P(t) is obtained from simulations by building a histogram of hbond lifetimes for each configuration, 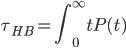 .
.
I did not find anywhere says the output frequency, I don't think the author print out every timestep, because my results are different from this paper. The method I used for hbond calculation is for each frame, I calculate all the hbonds, and push the hbond into live_array, if the hbonds existed in previous frame still exist in current frame, and push the hbond into dead_array if not. so the total number of appearance in live_array is total number of frames that this hbond is still alive, and the total number of appearance in dead_array is times this hbond had died. The number in live_array divided by number in dead_array is the average life time for that hbond. The simulation of water are running on a 40 box.
box.
**When plot this graph, set width=0.001, because 1step=0.002ps.
The average hbond lifetime is 0.0248 0.0131 ps, which is way too short, so what is the problem? look the picture below from reference 4.
0.0131 ps, which is way too short, so what is the problem? look the picture below from reference 4.
During simulation, some hbonds can incidentally go beyond the criterion for 10-20 fs(5-10 steps) , and some honds accidently formed due to duffusion motion. We need to rule out these two situations as much as possible.
there are two ways to improve this, i) change the hbond criteria, ii) develop factors that can adjust the break and reforming of hbond. let's first change the hbond criteria to Dist=3.5A, Ang=30.
After we change the criteria for hbond, the average hbond lifetime shift to about 0.0551 0.0323ps, this is still not enough, so we need to try method ii to calculate hbond lifetime. I defined a factor called "break", means if within $break time the hbond comes back, we still consider the hbond never died, and also add another factor called "continue", means if the hbond exist greater than $continue, then we count this as a true hbond.To realize this, I save all hbonds of one frame into hbond_live, but this time I also save the $frame, so hbonds_live={{donor acceptor hydrogen frame}...}, later on I will use this frame to decide the hbond lifetime.
0.0323ps, this is still not enough, so we need to try method ii to calculate hbond lifetime. I defined a factor called "break", means if within $break time the hbond comes back, we still consider the hbond never died, and also add another factor called "continue", means if the hbond exist greater than $continue, then we count this as a true hbond.To realize this, I save all hbonds of one frame into hbond_live, but this time I also save the $frame, so hbonds_live={{donor acceptor hydrogen frame}...}, later on I will use this frame to decide the hbond lifetime.
lifetime on top is output frequency=1, and break=1,5,10, continue=1,5,10, the average is 0.05ps, 0.06ps,0.10ps respectively .
lifetime at the bottom is output frequency=10, the average is 0.05, 0.57 and 0.89. Of cause, we can set the factor to be break=100, continue=100, or even longer, but the problem is if we we use bigger break or continue, we also need to increase sample time, from 1000 steps to 10000 or even longer, this will tremendously increase the calculation time. That is the reason some research step further to study correlation functions c(t), the probability that one hbond still bonded at time t provided the pair was bonded at t=0(independent of possible breaking in the interim time).
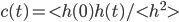
h(t)=1 if bonded, and 0 if not bonded
So P(t) is history dependent, but c(t) is history independent.
And also reactive flux, defined by derivative, which measure the effective decay rate of an initial set of hbonds.
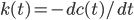
If we only need to care about hbond every 10 steps, when we run the simulation we don't need to spit out energy every step, so 10 steps would be a appropriate setting for energy output frequency, and this will also significantly decrease the calculation time.
To extract the new trajectory, we run command:
catdcd -o equi_298_10s.dcd -first 35001 -stride 10 equi_298.dcd
the output trajectory will have 20000/10=2000 frames.don't forget to change timestep * 0.02(from 0.002) in the *.tcl file when puts out time.
No surprise, after we load the trajectory extracted from original one, every 5 frames, and set break=1, continue=1, the average lifetime should be similar to above one. Now we try break=2, continue=2, the lifetime distribution will be like graph below.
The average lifetime is about 0.5ps~0.6ps, which is really close to the experiment and other people's simulation results.
reference 1:
The role of water in biology originated from hydrogen bond network. Hydrogen bond breaks and forms on fast time scale from tens of fs to ps. The experimental value of self diffusion coefficient of water at 298.2K is found by K Krynicki et al, to be 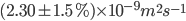 H-bond mean persistence time(~0.5ps). H-bond mean continuous lifetime(~0.2ps). Hydrogen bond has geometric(g) definition and energetic(e) definition.
H-bond mean persistence time(~0.5ps). H-bond mean continuous lifetime(~0.2ps). Hydrogen bond has geometric(g) definition and energetic(e) definition.
There are many ways to describe the distribution of hbond lifetime. C(t) is the most often calculated. It is the probability that the Hbond existing at the instant t=0 will also exist at the next instant t,regardless whether the bond was borken or not during the interval from 0 to t and whether or not it will exist after the instant t.
If the correlator C(t) is calculated only up to the moment the hbond breaking, then we obtain the H-bond survival probability. The probability of the fist Hbond breaking in time t after we have detected it at the moment t=0 will be denoted by 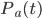 .
.
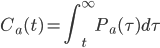
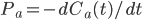
here is one link to a good description of hbonds in water.
http://www1.lsbu.ac.uk/water/water_hydrogen_bonding.html
References:
1.Real-Space Identification of Intermolecular Bonding with Atomic Force Microscopy
2.Fast and Slow Dynamics of Hydrogen Bonds in Liquid Water
3.Insights on Hydrogen-Bond Lifetimes in Liquid and Supercooled Water. H.F.M.C.Martiniano and N.Galamba.The Journal of Physical Chemistry.2013
4.Hydrogen bond lifetime distributions in computer-simulated water






