During the last two decades(History), molecular dynamics simulation has proved to be a paramount tool and was widely used to study protein structures, folding kinetics and thermodynamics, and structure-stability-function relationships. The MD simulation is still undergoing rapid developments in two general directions, one is toward developing new coarse-grained models and studying larger molecular systems while another direction is toward building high resolution models and explore more detailed and accurate picture of protein folding and asscociated processes.
Forces between different particles are derived from some analytical force model.
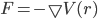
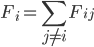
equations of motion are solved with a finite difference algorithm
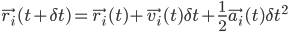
for all atom model:
People might think simulations are distinct from experiment, as they are predictions based on mathematical models, but I prefer it is just "numerical experiments". The equipments to do simulation are all kinds of softwares and packages, the coding of program or setting of parameters is not so different from learning the operation of HPLC, MS, NMR or whatever machines, and try to find the best condition for the measurement of samples.
The good thing about experiments is they are always true, what you need to do is to explain it. Simulations are skeptical in some way, but if the results are agree with experiment, then you'd better believe it, no matter what is inside the black box.
Both experiment and simulation takes times, shorter or longer depends on your skills. One pathetic thing about simulation is 90% of time they spend is on coding and debugging, 10% of time can be contributed to real thinking about the theory or method behind the simulation, be prepared to be frustrated, if you don't want to fiddling with your computer, then don't do simulations.
