Why do we need to run pH simulation?
Solution pH has a profound effect on the stability and the function of proteins by changing the protonation states of titratable groups. Although the importance of solution pH has long been recognized,molecular simulation techniques have traditionally neglected it.
In principle, the most rigorous way to esitimate an individual pKa value for a protein sidechaine would involve a free energy simulation connecting the protonated and deprotonated forms of the molecule:
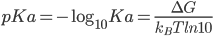
How do we define protonation/deprotonation?
"In the CPHMD method, protonation/deprotonation events are modeled by smoothly turning on/off electrostatic and van der Waals interactions involving titratable hydrogen atoms without breaking/forming bonds. Thus, titratable hydrogen atoms can be considered as dummy atoms that are covalently attached during the entire simulation time."
PMF function A* ( lambda - B) ^ 2
Calculation of pKa Values
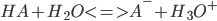
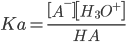
Henderson-Hasselbach equation:
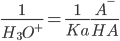
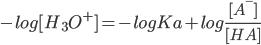
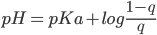
q is degree of protonation or occupancy:number of bound protons as a function of pH. The pKa of a titrating site is defined as the pH for which the site is 50% occupied:The pH for which the occupancy q is 0.5.
The pKa's of proteins were computed by fitting the unprotonated fractions(S) obtained from CPHMD simulations at various pH values to the generalized Henderson-Hasselbach(HH) equation:(1)
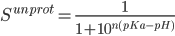
How do we get the titration curve of the whole protein.titration curve of a protein is a superposition of the curves for the individual types of groups
References:
1)Toward the accurate First-Principles Prediction of Ionization Equilibria in Proteins.
Here is a very useful link:
https://sites.google.com/site/cphmdtutorial/objective
And another tutorial:
http://www.mmtsb.org/workshops/mmtsb-ctbp_workshop_2009/Tutorials/CPHMD/cphmd_tutorial.html
http://www.farma.ku.dk/uploads/media/Lecture_3_NMR_titrations_2012.pdf